¶ Using R
General Information
R is a language and environment for statistical computing. It provides a wide variety of statistical and graphical techniques, and is highly flexible. This guide will show you how to run R in both graphical and command line modes.
¶ Using Rstudio
Rstudio is provided using a web front end called open ondemand. First you should login to the web interface here:
https://ondemand.wahab.hpc.odu.edu
You will login using your Midas account and password.
After logging in you will see this landing page:

In the top menu choose Interactive apps -> RStudio Server.
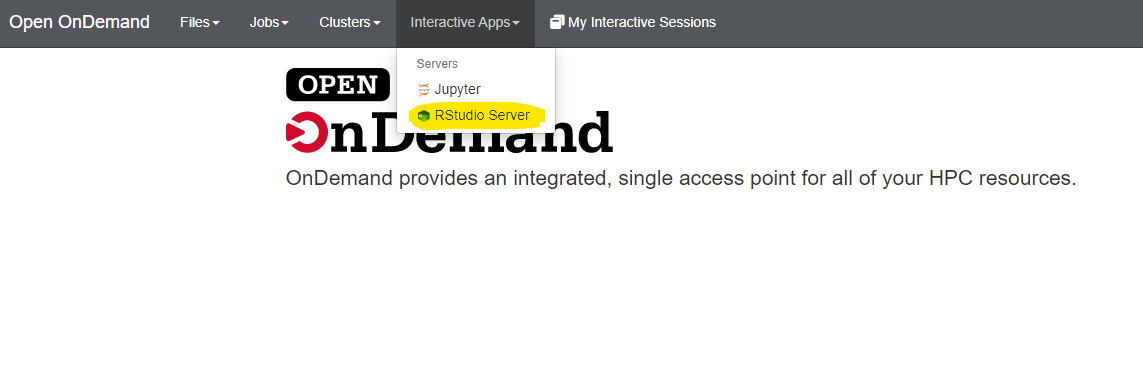
After choosing RStudio Server you will see the following window:
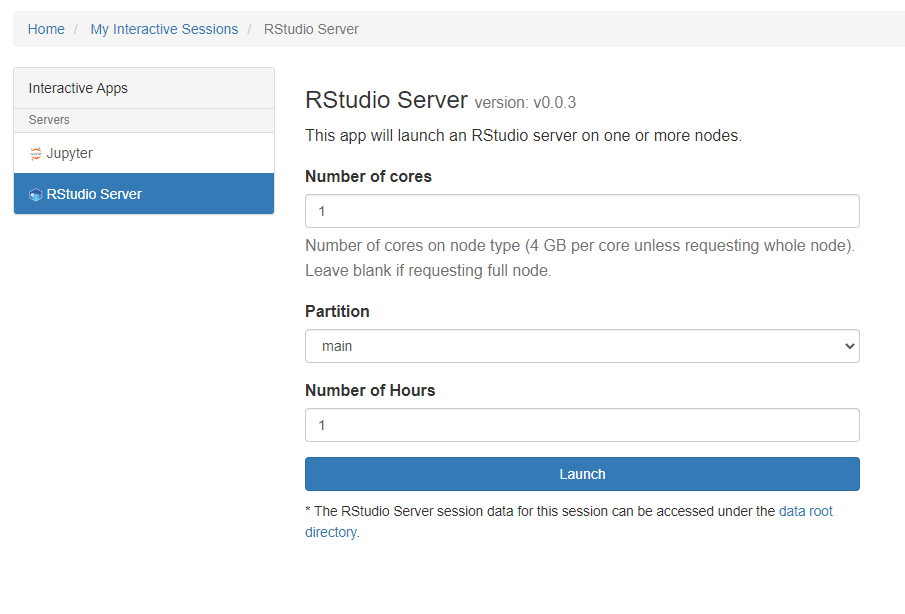
Please choose the number of cores you need and the duration of your session and click Launch. Please note the Number of Hours field as this is the time limit of your session. You can adjust this to a maximum of 24 hours. This will reserve time on one of the compute or GPU nodes in the cluster. When the session is ready for launch you will see this:
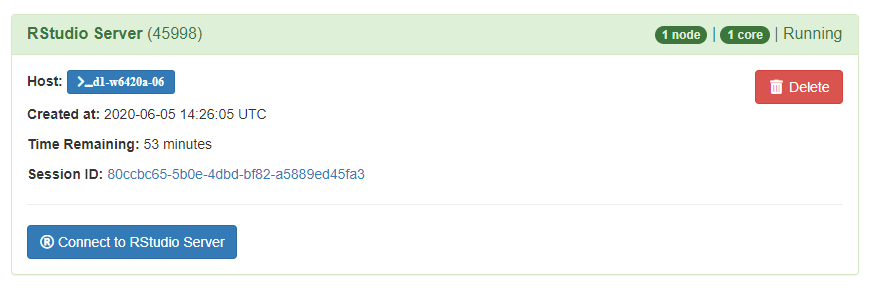
Click Connect to RStudio Server and your session with RStudio will be started. You will be placed into a new window with the RStudio session and you are ready to use the software. Please note that there is a time limit that can be adjusted on the length of your session.
¶ From the command line
In order to run R from the command line, you use the following commands.
First, you need to allocate a session on a compute node:
salloc
You will then use the following commands to load the R module. This loads the container environment and the required R singularity module.
enable_lmod
module load container_env R/4.3
You then have two options for running R:
¶ Running R interactively
In order to run R interactively you should run:
crun R
This will load the R program and you will have access to the R interface.
¶ Running an R script
You can also run R using a script. You can do this with the following command:
crun Rscript <script-name>
Just running crun Rscript will print the help information for Rscript.
¶ Submitting and R script to the cluster
You can submit an R script to the cluster using the SLURM scheduler.
Here is an example SLURM batch file:
#!/bin/bash
#SBATCH -J R_Job_Name
#SBATCH -c 40 # number of core requested, decrease to maximum of 32 on Turing
#SBATCH --mail-type=ALL
#SBATCH --mail-user=<email address>
enable_lmod
module load container_env R
crun Rscript <R script name>
Save this script to a file (ex. r-job.sh) and then submit to the scheduler:
sbatch r-job.sh
¶ Installing libraries
You can install R libraries into your home directory for use in R. In order to install libraries you should create a directory to store them in:
mkdir ~/Rpackages/
You can then install the desired package(s) into this directory using this command inside R:
install.packages("<package name>", lib="~/Rpackages/")
Finally you can point to the library location in your home directory to load the new package:
library(<package name>, lib.loc="~/Rpackages/")